About us
Learn how GA4GH helps expand responsible genomic data use to benefit human health.
Learn how GA4GH helps expand responsible genomic data use to benefit human health.
Our Strategic Road Map defines strategies, standards, and policy frameworks to support responsible global use of genomic and related health data.
Discover how a meeting of 50 leaders in genomics and medicine led to an alliance uniting more than 5,000 individuals and organisations to benefit human health.
GA4GH Inc. is a not-for-profit organisation that supports the global GA4GH community.
The GA4GH Council, consisting of the Executive Committee, Strategic Leadership Committee, and Product Steering Committee, guides our collaborative, globe-spanning alliance.
The Funders Forum brings together organisations that offer both financial support and strategic guidance.
The EDI Advisory Group responds to issues raised in the GA4GH community, finding equitable, inclusive ways to build products that benefit diverse groups.
Distributed across a number of Host Institutions, our staff team supports the mission and operations of GA4GH.
Curious who we are? Meet the people and organisations across six continents who make up GA4GH.
More than 500 organisations connected to genomics — in healthcare, research, patient advocacy, industry, and beyond — have signed onto the mission and vision of GA4GH as Organisational Members.
These core Organisational Members are genomic data initiatives that have committed resources to guide GA4GH work and pilot our products.
This subset of Organisational Members whose networks or infrastructure align with GA4GH priorities has made a long-term commitment to engaging with our community.
Local and national organisations assign experts to spend at least 30% of their time building GA4GH products.
Anyone working in genomics and related fields is invited to participate in our inclusive community by creating and using new products.
Wondering what GA4GH does? Learn how we find and overcome challenges to expanding responsible genomic data use for the benefit of human health.
Study Groups define needs. Participants survey the landscape of the genomics and health community and determine whether GA4GH can help.
Work Streams create products. Community members join together to develop technical standards, policy frameworks, and policy tools that overcome hurdles to international genomic data use.
GIF solves problems. Organisations in the forum pilot GA4GH products in real-world situations. Along the way, they troubleshoot products, suggest updates, and flag additional needs.
GIF Projects are community-led initiatives that put GA4GH products into practice in real-world scenarios.
The GIF AMA programme produces events and resources to address implementation questions and challenges.
NIF finds challenges and opportunities in genomics at a global scale. National programmes meet to share best practices, avoid incompatabilities, and help translate genomics into benefits for human health.
Communities of Interest find challenges and opportunities in areas such as rare disease, cancer, and infectious disease. Participants pinpoint real-world problems that would benefit from broad data use.
The Technical Alignment Subcommittee (TASC) supports harmonisation, interoperability, and technical alignment across GA4GH products.
Find out what’s happening with up to the minute meeting schedules for the GA4GH community.
See all our products — always free and open-source. Do you work on cloud genomics, data discovery, user access, data security or regulatory policy and ethics? Need to represent genomic, phenotypic, or clinical data? We’ve got a solution for you.
All GA4GH standards, frameworks, and tools follow the Product Development and Approval Process before being officially adopted.
Learn how other organisations have implemented GA4GH products to solve real-world problems.
Help us transform the future of genomic data use! See how GA4GH can benefit you — whether you’re using our products, writing our standards, subscribing to a newsletter, or more.
Join our community! Explore opportunities to participate in or lead GA4GH activities.
Help create new global standards and frameworks for responsible genomic data use.
Align your organisation with the GA4GH mission and vision.
Want to advance both your career and responsible genomic data sharing at the same time? See our open leadership opportunities.
Join our international team and help us advance genomic data use for the benefit of human health.
Discover current opportunities to engage with GA4GH. Share feedback on our products, apply for volunteer leadership roles, and contribute your expertise to shape the future of genomic data sharing.
Solve real problems by aligning your organisation with the world’s genomics standards. We offer software dvelopers both customisable and out-of-the-box solutions to help you get started.
Learn more about upcoming GA4GH events. See reports and recordings from our past events.
Speak directly to the global genomics and health community while supporting GA4GH strategy.
Be the first to hear about the latest GA4GH products, upcoming meetings, new initiatives, and more.
Questions? We would love to hear from you.
Read news, stories, and insights from the forefront of genomic and clinical data use.
Attend an upcoming GA4GH event, or view meeting reports from past events.
See new projects, updates, and calls for support from the Work Streams.
Read academic papers coauthored by GA4GH contributors.
Listen to our podcast OmicsXchange, featuring discussions from leaders in the world of genomics, health, and data sharing.
Check out our videos, then subscribe to our YouTube channel for more content.
View the latest GA4GH updates, Genomics and Health News, Implementation Notes, GDPR Briefs, and more.
Discover all things GA4GH: explore our news, events, videos, podcasts, announcements, publications, and newsletters.
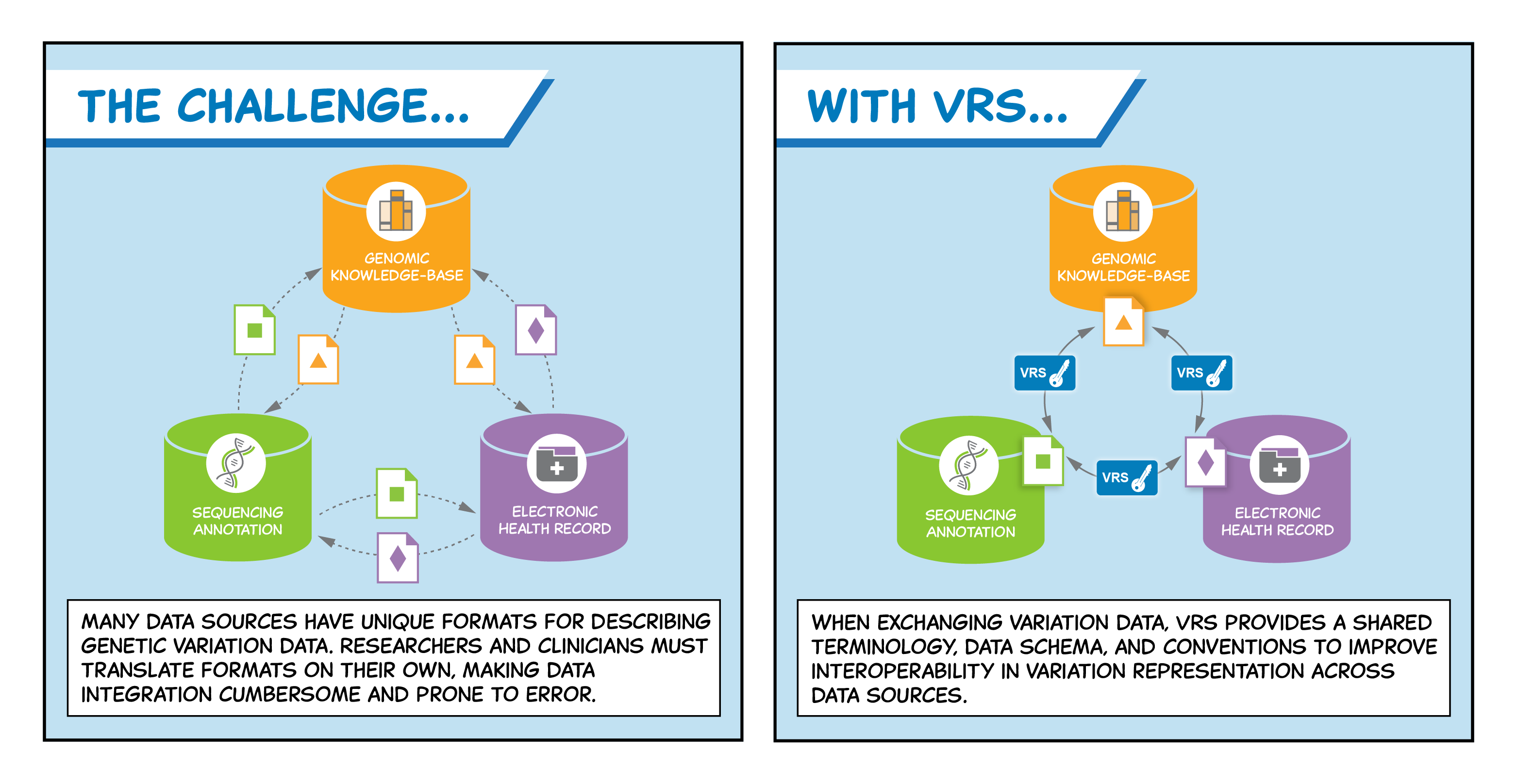
Between any two individuals, the human genome differs by around 0.1% — meaning each individual has millions of sites in their genome that collectively make them unique. The impact of genetic variation on human health and disease is complex and immense; in order to accelerate research and advance patient care, it is vital to unlock the potential of the world’s genetic variation data and exchange it reliably across diagnostic labs, electronic health records (EHRs), research institutions, and knowledge-bases. Developed by the GA4GH Genomic Knowledge Standards (GKS) Work Stream, the Variation Representation Specification (VRS) is a computational “language” that enables consistent communication across these systems.
Researchers, clinicians, and clinical laboratories
Dive deeper into this product!
Maximising the personal, public, research, and clinical value of genomic information will require clinicians, researchers, and testing laboratories to exchange genetic variation data reliably. VRS provides a community-developed specification to enable the precise representation and exchange of genetic variation data. The specification is designed to improve the reliability and utility of the clinical annotations that are central to personalised medicine, and consists of five components:
Catch up with all news and articles associated with Variation Representation Specification (VRS).