About us
Learn how GA4GH helps expand responsible genomic data use to benefit human health.
Learn how GA4GH helps expand responsible genomic data use to benefit human health.
Our Strategic Road Map defines strategies, standards, and policy frameworks to support responsible global use of genomic and related health data.
Discover how a meeting of 50 leaders in genomics and medicine led to an alliance uniting more than 5,000 individuals and organisations to benefit human health.
GA4GH Inc. is a not-for-profit organisation that supports the global GA4GH community.
The GA4GH Council, consisting of the Executive Committee, Strategic Leadership Committee, and Product Steering Committee, guides our collaborative, globe-spanning alliance.
The Funders Forum brings together organisations that offer both financial support and strategic guidance.
The EDI Advisory Group responds to issues raised in the GA4GH community, finding equitable, inclusive ways to build products that benefit diverse groups.
Distributed across a number of Host Institutions, our staff team supports the mission and operations of GA4GH.
Curious who we are? Meet the people and organisations across six continents who make up GA4GH.
More than 500 organisations connected to genomics — in healthcare, research, patient advocacy, industry, and beyond — have signed onto the mission and vision of GA4GH as Organisational Members.
These core Organisational Members are genomic data initiatives that have committed resources to guide GA4GH work and pilot our products.
This subset of Organisational Members whose networks or infrastructure align with GA4GH priorities has made a long-term commitment to engaging with our community.
Local and national organisations assign experts to spend at least 30% of their time building GA4GH products.
Anyone working in genomics and related fields is invited to participate in our inclusive community by creating and using new products.
Wondering what GA4GH does? Learn how we find and overcome challenges to expanding responsible genomic data use for the benefit of human health.
Study Groups define needs. Participants survey the landscape of the genomics and health community and determine whether GA4GH can help.
Work Streams create products. Community members join together to develop technical standards, policy frameworks, and policy tools that overcome hurdles to international genomic data use.
GIF solves problems. Organisations in the forum pilot GA4GH products in real-world situations. Along the way, they troubleshoot products, suggest updates, and flag additional needs.
GIF Projects are community-led initiatives that put GA4GH products into practice in real-world scenarios.
The GIF AMA programme produces events and resources to address implementation questions and challenges.
NIF finds challenges and opportunities in genomics at a global scale. National programmes meet to share best practices, avoid incompatabilities, and help translate genomics into benefits for human health.
Communities of Interest find challenges and opportunities in areas such as rare disease, cancer, and infectious disease. Participants pinpoint real-world problems that would benefit from broad data use.
The Technical Alignment Subcommittee (TASC) supports harmonisation, interoperability, and technical alignment across GA4GH products.
Find out what’s happening with up to the minute meeting schedules for the GA4GH community.
See all our products — always free and open-source. Do you work on cloud genomics, data discovery, user access, data security or regulatory policy and ethics? Need to represent genomic, phenotypic, or clinical data? We’ve got a solution for you.
All GA4GH standards, frameworks, and tools follow the Product Development and Approval Process before being officially adopted.
Learn how other organisations have implemented GA4GH products to solve real-world problems.
Help us transform the future of genomic data use! See how GA4GH can benefit you — whether you’re using our products, writing our standards, subscribing to a newsletter, or more.
Join our community! Explore opportunities to participate in or lead GA4GH activities.
Help create new global standards and frameworks for responsible genomic data use.
Align your organisation with the GA4GH mission and vision.
Want to advance both your career and responsible genomic data sharing at the same time? See our open leadership opportunities.
Join our international team and help us advance genomic data use for the benefit of human health.
Discover current opportunities to engage with GA4GH. Share feedback on our products, apply for volunteer leadership roles, and contribute your expertise to shape the future of genomic data sharing.
Solve real problems by aligning your organisation with the world’s genomics standards. We offer software dvelopers both customisable and out-of-the-box solutions to help you get started.
Learn more about upcoming GA4GH events. See reports and recordings from our past events.
Speak directly to the global genomics and health community while supporting GA4GH strategy.
Be the first to hear about the latest GA4GH products, upcoming meetings, new initiatives, and more.
Questions? We would love to hear from you.
Read news, stories, and insights from the forefront of genomic and clinical data use.
Publishes regular briefs exploring laws and regulations, including data protection laws, that impact genomic and related health data sharing
Translates findings from studies on public attitudes towards genomic data sharing into short blog posts, with a particular focus on policy implications
Attend an upcoming GA4GH event, or view meeting reports from past events.
See new projects, updates, and calls for support from the Work Streams.
Read academic papers coauthored by GA4GH contributors.
Listen to our podcast OmicsXchange, featuring discussions from leaders in the world of genomics, health, and data sharing.
Check out our videos, then subscribe to our YouTube channel for more content.
View the latest GA4GH updates, Genomics and Health News, Implementation Notes, GDPR Briefs, and more.
17 Jul 2023
Millions of genomes have been sequenced worldwide. The GA4GH standard refget quietly helps to decipher many of them. Now a new refget version opens up reliable reference genome retrieval to even more communities.
A widely-used tool that finds the exact references needed to pinpoint differences in our DNA just got a refresh.
On 17 July, the Standards Steering Committee of the Global Alliance for Genomics and Health (GA4GH) voted to release refget v2.0. With better compatibility for a range of reference genome names, formats, and systems, the new version of refget makes it easier than ever to retrieve verified genomic reference sequences.
A vital infrastructure
You may not even realise that you’re using refget already.
“Almost anyone who uses a CRAM file uses refget,” said Timothe Cezard, co-leader of the team that produced the new refget version and a project lead at EMBL’s European Bioinformatics Institute (EMBL-EBI). “Compression, decompression, all CRAM tools — say, for conversion to other formats or direct analysis — sit on top of refget.”
CRAM is a popular and efficient file format for storing DNA sequences, able to reduce storage costs by up to 50%. It achieves that staggering compression by relying on a reference sequence — a bit of DNA considered typical. Compare your own genes against the reference, and you will begin to see variation: genetic differences that could lead to everything from freckles to a high risk of breast cancer.
Instead of storing all three billion base pairs of the reference sequence alongside the DNA being studied, CRAM files simply hold onto the reference sequence’s name.
When it’s time to decompress the data, refget steps in — helping you “get” the “reference” you need.
Solving the dictionary dilemma
CRAM is just one example of how refget removes dangerous uncertainty from genomic data.
“refget identifies each reference sequence using its inherent unique qualities, so you can always trust that a sequence contains what it says on the label,” said Andrew Yates, a founding developer of refget and a team leader at EMBL-EBI. “The consequences of comparing genomic data to incorrect or misaligned reference sequences are serious. Genetic variants may be classified as pathogenic or harmless incorrectly, and patients could receive improper care. Being exact matters.”
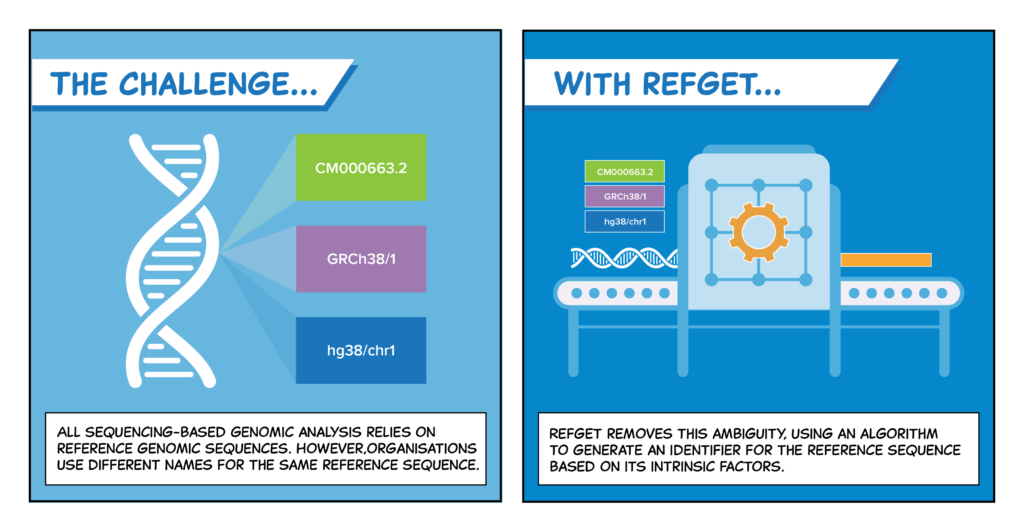
By assigning a unique identifier to reference sequences, refget solves a tricky naming problem in genomics.
Central authorities like the International Nucleotide Sequence Database Collaboration (INSDC), Ensembl, and the University of California, Santa Cruz (UCSC) Genome Browser use different naming conventions for the same reference sequence.
Think of how the Oxford English Dictionary and Merriam-Webster sometimes spell and define the same English word differently. Then try to convince a British English speaker to use “color” instead of “colour,” and you will see the challenge of standardising nomenclature.
Non-unique names create even more uncertainty when analysing data. For instance, another common naming convention counts by chromosome number, starting with chromosome 1 as the largest. But the reference genomes of many organisms have a chromosome called “1.” How do you know you’re getting a human chromosome and not a mouse, for instance? Which “1” is the right one to use?
refget clears up any confusion.
“refget is very straightforward. You’ve got a name, you grab a sequence. You have a sequence, you construct the name,” said Cezard. “You don’t need to rely on any naming authority.”
Why you need refget for any genomic analysis
For the initial refget release in 2018, the GA4GH Large Scale Genomics Work Stream tailored the API to support CRAM.
But Yates and team quickly realised that refget could smooth over issues in other genomic data formats and models. VCF and SAM also support refget identifiers, with growing community interest in using them.
“refget is a fundamental building block for GA4GH standards,” said Yates. “It can solve problems beyond CRAM, for any file format or data model that requires a reference sequence. With refget, you know exactly what sequence you’re talking about.”
For instance, refget is already solving problems for the GA4GH Variation Representation Specification (VRS), which provides a framework for describing genetic variants that computers can easily compare and analyse.
Yates and Cezard worked closely with the VRS team to develop refget v2.0, which supports VRS sequence identifiers. Now hospitals, laboratories, and databases like NIH-funded ClinGen, which use VRS to represent and share genetic variants, link to reference sequences via refget.
“In large part due to refget, GA4GH VRS makes sharing and comparing variant data across institutions much more reliable. refget lets us pinpoint the exact reference sequence, which then helps us represent the variation unambiguously,” said Larry Babb, a leader of the VRS team, principal software engineer at the Broad Institute of MIT and Harvard, and a GA4GH Driver Project Champion for ClinGen.
“By using refget identifiers in VRS, we can tackle important interoperability challenges that arise when comparing evidence from new reference sequences. The strategy has already worked in the real world in a project with the Atlas of Variant Effects Alliance,” said Alex Wagner, the other VRS team leader, who is a principal investigator at Nationwide Children’s Hospital and GA4GH Driver Project Champion for the Variant Interpretation for Cancer Consortium.
Another major resource for the genomics community, the European Nucleotide Archive (ENA), has already implemented refget v2.0.
ENA contains all sequenced DNA and RNA in the public domain — nearly three billion sequences. To decompress files from the database, researchers use the CRAM reference registry, which runs on refget.
The new version of refget will also debut in the Ensembl genome browser. This collection of more than 50,000 genomes (representing great diversity within and among species, from humans to maize to zebrafish) offers tools for analysis and comparison.
“refget is powering our new Ensembl infrastructure. These refget endpoints will be made available in the near future and will provide access to Ensembl hosted protein and transcript sequences,” said Yates.
New features in v2.0
The latest version of refget expands the capabilities of the API, making it more accessible and compatible with other systems.
Work with the VRS team led to a new preferred algorithm for defining identifiers. Other new features detailed in the specification include recommended best practices (such as lowercase naming authority strings), and options when searching for a specific identifier (with or without a namespace).
One key change — aimed at expanding the groups who can benefit from refget — allows you to search not just by unique refget identifier, but by another naming convention. In dictionary terms, you can look up either “colour” or “color” and still retrieve the right definition.
“refget servers can now retrieve the same sequence using a different naming convention. The new version is interoperable with other systems that rely on naming authorities, so you can search even if you don’t have access to the reference sequence itself,” said Cezard.
“You can enter a name that isn’t a refget identifier and still get the same verified, reliable sequence — which you can then recompute into a refget identifier,” he added.
The new version includes technical solutions for handling non-unique names.
These major v2.0 upgrades don’t entail major work for implementers: all existing refget clients can continue using the API. The only breaking change is a minimal one and makes refget servers compatible with the GA4GH Service Info API, which helps find web services for analysing genomic data.
refget for an entire genome
Building on the same principles as refget, the team is currently developing a new specification that verifies the identity of collections of sequences.
“refget defines a name for a single sequence, like a chromosome. Sequence Collections defines a name for a group of sequences, which we would often use for assemblies or a whole genome,” said Cezard.
Sequence Collections will offer many new features beyond defining names, including searching within and comparing collections.
In the meantime, Cezard, Yates, and collaborators aim to strengthen support for refget in a wide range of genomic file formats, from BED to SAM to VCF, and demonstrate how useful refget can be.
“refget already safeguards a crucial step in genomic analysis for researchers around the world,” said Yates. “This second version reinforces how vital the concept of unique identifiers really is, whether you are identifying a single reference sequence, an entire genome, or even a pangenome.”
