About us
Learn how GA4GH helps expand responsible genomic data use to benefit human health.
Learn how GA4GH helps expand responsible genomic data use to benefit human health.
Our Strategic Road Map defines strategies, standards, and policy frameworks to support responsible global use of genomic and related health data.
Discover how a meeting of 50 leaders in genomics and medicine led to an alliance uniting more than 5,000 individuals and organisations to benefit human health.
GA4GH Inc. is a not-for-profit organisation that supports the global GA4GH community.
The GA4GH Council, consisting of the Executive Committee, Strategic Leadership Committee, and Product Steering Committee, guides our collaborative, globe-spanning alliance.
The Funders Forum brings together organisations that offer both financial support and strategic guidance.
The EDI Advisory Group responds to issues raised in the GA4GH community, finding equitable, inclusive ways to build products that benefit diverse groups.
Distributed across a number of Host Institutions, our staff team supports the mission and operations of GA4GH.
Curious who we are? Meet the people and organisations across six continents who make up GA4GH.
More than 500 organisations connected to genomics — in healthcare, research, patient advocacy, industry, and beyond — have signed onto the mission and vision of GA4GH as Organisational Members.
These core Organisational Members are genomic data initiatives that have committed resources to guide GA4GH work and pilot our products.
This subset of Organisational Members whose networks or infrastructure align with GA4GH priorities has made a long-term commitment to engaging with our community.
Local and national organisations assign experts to spend at least 30% of their time building GA4GH products.
Anyone working in genomics and related fields is invited to participate in our inclusive community by creating and using new products.
Wondering what GA4GH does? Learn how we find and overcome challenges to expanding responsible genomic data use for the benefit of human health.
Study Groups define needs. Participants survey the landscape of the genomics and health community and determine whether GA4GH can help.
Work Streams create products. Community members join together to develop technical standards, policy frameworks, and policy tools that overcome hurdles to international genomic data use.
GIF solves problems. Organisations in the forum pilot GA4GH products in real-world situations. Along the way, they troubleshoot products, suggest updates, and flag additional needs.
GIF Projects are community-led initiatives that put GA4GH products into practice in real-world scenarios.
The GIF AMA programme produces events and resources to address implementation questions and challenges.
NIF finds challenges and opportunities in genomics at a global scale. National programmes meet to share best practices, avoid incompatabilities, and help translate genomics into benefits for human health.
Communities of Interest find challenges and opportunities in areas such as rare disease, cancer, and infectious disease. Participants pinpoint real-world problems that would benefit from broad data use.
The Technical Alignment Subcommittee (TASC) supports harmonisation, interoperability, and technical alignment across GA4GH products.
Find out what’s happening with up to the minute meeting schedules for the GA4GH community.
See all our products — always free and open-source. Do you work on cloud genomics, data discovery, user access, data security or regulatory policy and ethics? Need to represent genomic, phenotypic, or clinical data? We’ve got a solution for you.
All GA4GH standards, frameworks, and tools follow the Product Development and Approval Process before being officially adopted.
Learn how other organisations have implemented GA4GH products to solve real-world problems.
Help us transform the future of genomic data use! See how GA4GH can benefit you — whether you’re using our products, writing our standards, subscribing to a newsletter, or more.
Join our community! Explore opportunities to participate in or lead GA4GH activities.
Help create new global standards and frameworks for responsible genomic data use.
Align your organisation with the GA4GH mission and vision.
Want to advance both your career and responsible genomic data sharing at the same time? See our open leadership opportunities.
Join our international team and help us advance genomic data use for the benefit of human health.
Discover current opportunities to engage with GA4GH. Share feedback on our products, apply for volunteer leadership roles, and contribute your expertise to shape the future of genomic data sharing.
Solve real problems by aligning your organisation with the world’s genomics standards. We offer software dvelopers both customisable and out-of-the-box solutions to help you get started.
Learn more about upcoming GA4GH events. See reports and recordings from our past events.
Speak directly to the global genomics and health community while supporting GA4GH strategy.
Be the first to hear about the latest GA4GH products, upcoming meetings, new initiatives, and more.
Questions? We would love to hear from you.
Read news, stories, and insights from the forefront of genomic and clinical data use.
Publishes regular briefs exploring laws and regulations, including data protection laws, that impact genomic and related health data sharing
Translates findings from studies on public attitudes towards genomic data sharing into short blog posts, with a particular focus on policy implications
Attend an upcoming GA4GH event, or view meeting reports from past events.
See new projects, updates, and calls for support from the Work Streams.
Read academic papers coauthored by GA4GH contributors.
Listen to our podcast OmicsXchange, featuring discussions from leaders in the world of genomics, health, and data sharing.
Check out our videos, then subscribe to our YouTube channel for more content.
View the latest GA4GH updates, Genomics and Health News, Implementation Notes, GDPR Briefs, and more.
17 Jun 2022
The latest GDPR Brief, written by Adrian Thorogood, considers the data protection implications of publishing metadata to enable discovery.
Metadata is data that describes data or another resource (e.g., a biospecimen), and is a key part of what ensures data are findable, accessible, interoperable and re-useable (FAIR). Metadata may in some circumstances be considered personal data – sometimes to the surprise of organizations seeking to process or publish them. What counts as metadata, what types of metadata are needed to realize FAIRness, and whether or not metadata need to be published depend on the context.
Some metadata models describe datasets (i.e., collections or cohorts) as part of data catalogues. Data catalogues are typically published to enable the research community to discover relevant datasets for their projects. Dataset metadata may include the following:
Organizations publishing dataset metadata should keep a look out for certain risks of privacy breach. For example, some administrative metadata is personal data–such as the names, email addresses, and affiliations of the responsible Principal Investigators. Permission may be required to publish these data. Statistical descriptions of datasets may leak personal data, e.g., where the datasets are small (e.g., in rare disease contexts) or incrementally updated. Indeed, where a small number of individuals are added to a dataset, the difference between the new statistical description and the old one may leak personal data. Publishing genomic summary statistics might also be subject to attacks that reveal if an individual is a member of the dataset (in turn potentially revealing sensitive attributes like disease status). Keep in mind also that generating statistics from underlying health and genetic data is a form of processing that requires adherence to applicable data protection regulations.
Bioinformaticians also commonly use the term metadata for data that describes individual research participants (e.g., donor ID, age, gender, primary phenotype), as well as samples extracted from an individual (e.g., tissue type, preparation), and sequencing experiments run on these samples. Individual-level metadata enable the comparison, linkage, and re-use of sequence data. A handful of individual-level metadata fields may also be published to facilitate data discovery, such as each subject’s age, gender, and phenotype. The label “metadata” should not, however, distract from the fact that these data are in essence individual-level demographic and health data (albeit usually low-dimensional data).
As with any human data, controllers must assess the identifiability of metadata before proceeding with processing or publication. In doing so they must consider all means reasonably likely to be used by any person to re-identify the individual and objective factors including the costs and time required, available technology at the time of the processing and technological developments. A contextual, risk-based approach is recommended for identifiability assessments. The need to enable discovery by the research community offers a justification for publishing metadata, though publication also limits available safeguards and reduces contextual certainty over identifiability. Aggregate data or a handful of individual-level data fields are typically sufficient to enable data discovery. A data protection impact assessment can be performed to demonstrate that such data poses lower risks of both re-identification and disclosure of sensitive information than the underlying data. It is often possible to mathematically demonstrate a low or nil risk of re-identification for such data using frameworks like k-anonymity. Release of metadata can also be controlled through a Beacon-type query interface, limiting the amount of metadata released to any particular requestor. Differential privacy controls can be added to these interfaces that protect against a requestor making multiple different queries in the aim of re-identification. Finally, layered approaches can be adopted to balance discovery and protection, e.g., by releasing a subset of metadata publicly, while limiting access to richer metadata to registered requesters.
Data archives can also play a role in balancing discovery and protection. Data submission agreements should clarify who is responsible for ensuring the anonymity of metadata that will be published in a data catalogue. Data protection by design can inform the structure of metadata models. Metadata field descriptions should be clear and focused, and potentially constrained by controlled vocabularies, to avoid the risk of inadvertent entry and release of personal data. Data models can also intuitively segregate sensitive (e.g., individual disease status) and non-sensitive (e.g., sample type) attributes (see Figure 1).
From a policy perspective, it is important not to hyperfocus on the data protection risks of processing or publishing metadata. Metadata not only facilitates scientific activity; it also supports both data protection and sustainability. Ecosystems where researchers can effectively discover, access, and re-use relevant genomic and health data are both more ethical – avoiding unnecessary disclosures of irrelevant data – and more efficient – saving both researchers and data providers time and resources discovering, accessing, and re-using data.
Figure 1. Segregation of sensitive and non-sensitive metadata
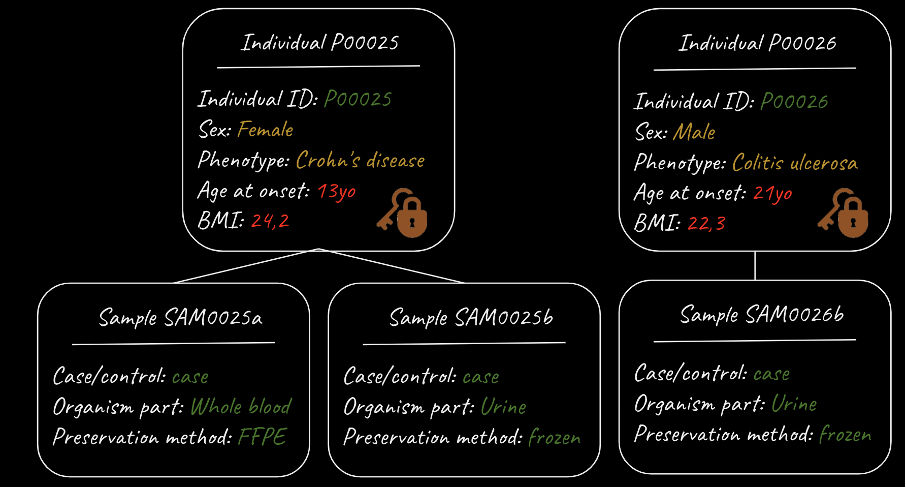
This figure shows a metadata model with fields describing individuals, as well as additional fields describing one or more samples taken from a given individual. The metadata model is nested – each sample can equally be described by the associated individual metadata. The metadata model also segregates “sensitive” individual level metadata (which is kept secure), from non-sensitive sample-level metadata. The figure is provided courtesy of Coline Thomas, European Molecular Biology Laboratory – European Bioinformatics Institute (EMBL-EBI).
Adrian Thorogood is a legal researcher at the Luxembourg Centre for Systems Biomedicine, University of Luxembourg.
See all previous briefs.
Please note that GDPR Briefs neither constitute nor should be relied upon as legal advice. Briefs represent a consensus position among Forum Members regarding the current understanding of the GDPR and its implications for genomic and health-related research. As such, they are no substitute for legal advice from a licensed practitioner in your jurisdiction.
