About us
Learn how GA4GH helps expand responsible genomic data use to benefit human health.
Learn how GA4GH helps expand responsible genomic data use to benefit human health.
Our Strategic Road Map defines strategies, standards, and policy frameworks to support responsible global use of genomic and related health data.
Discover how a meeting of 50 leaders in genomics and medicine led to an alliance uniting more than 5,000 individuals and organisations to benefit human health.
GA4GH Inc. is a not-for-profit organisation that supports the global GA4GH community.
The GA4GH Council, consisting of the Executive Committee, Strategic Leadership Committee, and Product Steering Committee, guides our collaborative, globe-spanning alliance.
The Funders Forum brings together organisations that offer both financial support and strategic guidance.
The EDI Advisory Group responds to issues raised in the GA4GH community, finding equitable, inclusive ways to build products that benefit diverse groups.
Distributed across a number of Host Institutions, our staff team supports the mission and operations of GA4GH.
Curious who we are? Meet the people and organisations across six continents who make up GA4GH.
More than 500 organisations connected to genomics — in healthcare, research, patient advocacy, industry, and beyond — have signed onto the mission and vision of GA4GH as Organisational Members.
These core Organisational Members are genomic data initiatives that have committed resources to guide GA4GH work and pilot our products.
This subset of Organisational Members whose networks or infrastructure align with GA4GH priorities has made a long-term commitment to engaging with our community.
Local and national organisations assign experts to spend at least 30% of their time building GA4GH products.
Anyone working in genomics and related fields is invited to participate in our inclusive community by creating and using new products.
Wondering what GA4GH does? Learn how we find and overcome challenges to expanding responsible genomic data use for the benefit of human health.
Study Groups define needs. Participants survey the landscape of the genomics and health community and determine whether GA4GH can help.
Work Streams create products. Community members join together to develop technical standards, policy frameworks, and policy tools that overcome hurdles to international genomic data use.
GIF solves problems. Organisations in the forum pilot GA4GH products in real-world situations. Along the way, they troubleshoot products, suggest updates, and flag additional needs.
GIF Projects are community-led initiatives that put GA4GH products into practice in real-world scenarios.
The GIF AMA programme produces events and resources to address implementation questions and challenges.
NIF finds challenges and opportunities in genomics at a global scale. National programmes meet to share best practices, avoid incompatabilities, and help translate genomics into benefits for human health.
Communities of Interest find challenges and opportunities in areas such as rare disease, cancer, and infectious disease. Participants pinpoint real-world problems that would benefit from broad data use.
The Technical Alignment Subcommittee (TASC) supports harmonisation, interoperability, and technical alignment across GA4GH products.
Find out what’s happening with up to the minute meeting schedules for the GA4GH community.
See all our products — always free and open-source. Do you work on cloud genomics, data discovery, user access, data security or regulatory policy and ethics? Need to represent genomic, phenotypic, or clinical data? We’ve got a solution for you.
All GA4GH standards, frameworks, and tools follow the Product Development and Approval Process before being officially adopted.
Learn how other organisations have implemented GA4GH products to solve real-world problems.
Help us transform the future of genomic data use! See how GA4GH can benefit you — whether you’re using our products, writing our standards, subscribing to a newsletter, or more.
Join our community! Explore opportunities to participate in or lead GA4GH activities.
Help create new global standards and frameworks for responsible genomic data use.
Align your organisation with the GA4GH mission and vision.
Want to advance both your career and responsible genomic data sharing at the same time? See our open leadership opportunities.
Join our international team and help us advance genomic data use for the benefit of human health.
Discover current opportunities to engage with GA4GH. Share feedback on our products, apply for volunteer leadership roles, and contribute your expertise to shape the future of genomic data sharing.
Solve real problems by aligning your organisation with the world’s genomics standards. We offer software dvelopers both customisable and out-of-the-box solutions to help you get started.
Learn more about upcoming GA4GH events. See reports and recordings from our past events.
Speak directly to the global genomics and health community while supporting GA4GH strategy.
Be the first to hear about the latest GA4GH products, upcoming meetings, new initiatives, and more.
Questions? We would love to hear from you.
Read news, stories, and insights from the forefront of genomic and clinical data use.
Publishes regular briefs exploring laws and regulations, including data protection laws, that impact genomic and related health data sharing
Translates findings from studies on public attitudes towards genomic data sharing into short blog posts, with a particular focus on policy implications
Attend an upcoming GA4GH event, or view meeting reports from past events.
See new projects, updates, and calls for support from the Work Streams.
Read academic papers coauthored by GA4GH contributors.
Listen to our podcast OmicsXchange, featuring discussions from leaders in the world of genomics, health, and data sharing.
Check out our videos, then subscribe to our YouTube channel for more content.
View the latest GA4GH updates, Genomics and Health News, Implementation Notes, GDPR Briefs, and more.
16 Nov 2023
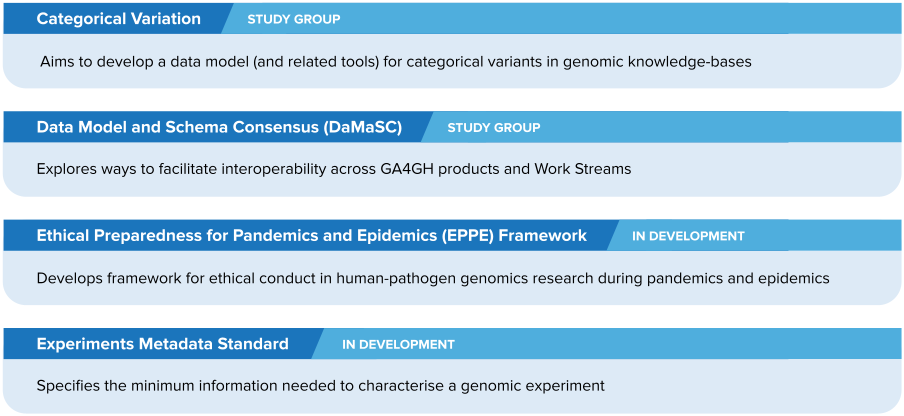
Anyone interested is invited to join the Categorical Variation Study Group, the Data Model and Schema Consensus (DaMaSC) Study Group, and groups developing the Experiments Metadata Standard and Ethical Preparedness for Pandemics and Epidemics Framework.

How do you figure out if your CRAM file came from a single cell assay or bulk sequencing experiment?
Have you noticed that key evidence linking genes and disease is often attached to broad categories of variants, which are hard to find and compare?
In the next pandemic, how do we make it easier for human-pathogen genomics researchers to speed up their work while maintaining the same level of ethical conduct?
And why doesn’t every GA4GH Work Stream represent data using a shared language?
Four new groups at the Global Alliance for Genomics and Health (GA4GH) will address those questions, and more.
Sign up to join the conversation if you want to shape the way we assemble genomic data, use data to fight disease (in a single patient all the way up to a pandemic), and break down silos that make data ineffective.
The four groups are:
Learn more about each group below.
Two groups have begun building products — the EPPE Framework and the Experiments Metadata Standard — after the GA4GH Standards Steering Committee voted in September 2023 to green-light their development.
The other two are Study Groups: teams that survey the landscape of the genomics and health community, define needs, and determine whether a GA4GH product could help.
Anyone in the genomics and health community is welcome to participate in the groups. You will work alongside contributors from around the world to explore and build GA4GH products.
Researchers often obtain genomic data as CRAM or VCF documents, but these contain little information on the experiment that produced the results. For example: are the data from whole genome sequencing, transcriptomics, or another kind of epigenomic experiment?
That lack of clarity is an issue.
“It is important to get details about the experiment that produced the genomic data you are dealing with,” said David Bujold, Lead of the standard development team and Bioinformatics Manager at McGill University.
“Instruments and sequencing techniques will affect the results. You need to understand the purpose of an experiment, like whether it targeted only certain regions of the genome, or a specific epigenetic feature, which will lead you to interpret the data in different ways. The GA4GH Experiments Metadata Standard aims to help clear up these ambiguities,” Bujold said.
The team, which operates within the GA4GH Discovery Work Stream, has begun developing a checklist of the minimum experimental information that should be captured about every high-throughput sequencing assay.
“We have seen time and again that when researchers move quickly during a global health emergency, they have sometimes completely disregarded basic ethical principles. And yet, speed is of the essence to get research results into the hands of decision-makers,” said Anja Bedeker, EPPE Lead, Co-Chair of the Public Health Alliance for Genomic Epidemiology (PHA4GE), and a Research Associate at the South African National Bioinformatics Institute.
“The GA4GH Ethical Preparedness for Pandemics and Epidemics Framework will present a path for balancing these two crucial obligations in human-pathogen genomics research,” Bedeker said.
The EPPE Framework will be a practical tool that researchers can use when studying human-pathogen genomics during pandemics and epidemics.
To build the new framework, the GA4GH Regulatory & Ethics Work Stream is collaborating with the PHA4GE Ethics and Data Sharing Working Group.
Say you’re reading a study on breast cancer, and you see the phrase “TP53 R248 mutations” (Berns et al. 1998). This statement doesn’t refer to a single change on a single gene. It refers to all possible variants that lead to changes at the amino acid position R248 on the protein TP53.
The phrase “TP53 R248 mutations” is an example of a categorical variant — a way for researchers to share knowledge about an entire category of genomic variation in one statement. Knowledge-bases like CIViC, ClinVar, OncoKB, and the JaxCKB use categorical variants to describe genomic evidence.
The problem? When a patient or research participant comes in with a very specific variant, you have to manually search many of these categorical variants to find the crucial evidence you need — whether or not that particular person’s variant leads to disease.
“In the GA4GH Categorical Variation Study Group, we’re going to tackle the complexity and confusion of categorical variants head-on. We hope to build a data model that helps researchers quickly find and connect categorical variants — and thus get better answers for patients and research participants,” said Daniel Puthawala, CatVar Lead and a Postdoctoral Scientist at Nationwide Children’s Hospital.
For many reasons — political, ethical, commercial, circumstantial, and otherwise — biomedical data often get stuck in silos. GA4GH was founded to expand responsible use of data in order to benefit human health. The new DaMaSC Study Group will help make that mission a reality by ramping up interoperability in the way GA4GH products represent data.
“GA4GH standards power data search and analysis worldwide. So we are deeply invested in ensuring standards work together to disassemble silos, rather than reinforce them accidentally,” said Kathy Reinold, an independent consultant who leads DaMaSC. “The GA4GH Data Model and Schema Consensus Study Group is exploring how to create an easy starting point for representing genomic and related data in an interoperable way.”
The team wants to establish a resource for anyone creating new genomic data representations, or schemas. DaMaSC members will study what conventions, guidelines, and existing schemas can simplify data representation. The Study Group also plans to explore what a common vocabulary of data representation across Work Streams would look like — in service of making GA4GH products easier to use and combine.